For example, multiple restingstate networks can be revealed from a single rsfMRI study without the need to administer one or more prescribed behavioral tasks, typically by measuring BOLD signal correlations relative to a "seed" region of interest, or by using multivariate component models to identify networks based on statistical criteriaCalculate group average of Fisher z scores 4Perform two sample ttest of group averagesRestingState fMRI rsfMRI is a method aimed at examining intrinsic networks in the brain while no task is performed (rest);

Functional Connectomics From Resting State Fmri Trends In Cognitive Sciences
Seed region fmri
Seed region fmri-The middle panel shows a timeseries extracted from a seed region The bottom panel shows the interaction regressor, created by multiplying the above regressors Note how the 1's, when multiplied by the seed timeseries, invert the sign of the timeseries if the timeseries is going down, in the interaction regressor it will go upFunctional magnetic resonance imaging (fMRI) has become a powerful tool to study brain dynamics with relatively fine spatial resolution One popular paradigm, given the indirect nature of the method, is the block design, alternating task blocks with blocks of passive rest Voxels showing a correlation with the seed region above a certain



Behavioral Sciences Free Full Text Random Forest Classification Of Alcohol Use Disorder Using Eeg Source Functional Connectivity Neuropsychological Functioning And Impulsivity Measures Html
1 vector X ij and the outcome variables to be Y qij where q = 1, ,Q denotes the seed region (Y 1 ij) and its (Q − 1) regions of interest (ROIs), i = 1, , n denotes the ith subject, and j = 1, , J denotes the jth fMRI measurementSeedbased connectivity metrics characterize the connectivity patterns with a predefined seed or ROI (Region of Interest) These metrics are often used when researchers are interested in one, or a few, individual regions and would like to analyze in detail the connectivity patterns between these areas and the rest of the brainOr using multivariate methods, such
However, using the same rsfMRI data, with the seed region defined in the canonical counterpart of the Broca's area according to the standard coordinates, the seedbased FC mapping (Fig 4E,FResting state fMRI analysis using seed based and ICA methods Abstract In this paper, we analyze the functional connectivity between the various parts of the brain using resting state fMRI(Functional Magnetic Resonance Imaging)18 by American Journal of Neuroradiology
Similar to conventional taskrelated fMRI, the BOLD fMRI signal is measured throughout the experiment (panel a) Conventional taskdependent fMRI can be used to select a seed region of interest (panel b) To examine the level of functional connectivity between the selected seed voxel i and a second brain region j (for example a region in theOr using multivariate methods, such as independent component analysis (ICA)This correlation between timeseries of different regions of the brain Restingstate functional connectivity



Figure 5 From Multimodal Imaging In Alzheimer S Disease Validity And Usefulness For Early Detection Semantic Scholar



The Role Of Resting State Functional Mri For Clinical Preoperative Language Mapping Cancer Imaging Full Text
To better understand intrinsic brain connections in major depression, we used a neuroimaging technique that measures resting state functional connectivity using functional MRI (fMRI) Three different brain networks—the cognitive control network, default mode network, and affective network—were investigated Compared with controls, in depressed subjects each ofIn investigations of the brain's resting state using functional magnetic resonance imaging (fMRI), a seedbased approach is commonly used to identify brain regions that are functionally connected The seed is typically identified based on anatomical landmarks, coordinates, or the location of brain activity during a separate taskFMRI, during which the same subjects performed bilateral finger tapping After determining the correlation between the BOLD time course of the seed region and that of all other areas in the brain, the authors found that the left somatosensory cortex was highly correlated with homologous areas in the contralateral hemisphere




Seed Regions Of The Seed Based Resting State Analysis Seed Regions And Download Scientific Diagram



Fmri Connectivity Analysis In Afni Ppt Video Online Download
The map represents a restingstate functional connectivity analysis performed on 1,000 human subjects, with the seed placed at the currently selected location Thus, it displays brain regions that are coactivated across the restingstate fMRI time series with the seedAfter defining a seed mask in the Montreal Neurological Institute (MNI) standard space (Fig 1a), the time course of each voxel within the seed region was extracted from the rsfMRI datasets (Fig 1b), serving as the data matrix in sparse representation Importantly, both the wholebrain functional connectivity patterns and the local timevaryingFunctionally, the brain is subspecialized for perceptual and



Multimodal Neuroimaging Of Prosody Applied Social Neuroscience And Perception Laboratory



Onlinelibrary Wiley Com Doi Pdf 10 1002 Mrm
Summarize regions by average time course 2For each subject, calculate correlations between seed region and other regions of interest 3Transform Pearson correlations by Fisher's z;Recent research into restingstate functional magnetic resonance imaging (fMRI) has shown that the brain is very active during rest patterns can be identified based on how similar the time profile is to a seed region's time profile However, this method requires a seed region and can only identify one resting state network at a timeRestingstate fMRI The brain controls all the complex functions in the human body Structurally, the brain is organized grossly into different regions specialized for processing and relaying neural signals;




Does Resting State Functional Connectivity Differ Between Mild Cognitive Impairment And Early Alzheimer S Dementia Journal Of The Neurological Sciences



Fsl Fmri Resting State Seed Based Connectivity Neuroimaging Core 0 1 1 Documentation
The seed sequence is essential for the binding of the miRNA to the mRNA The seed sequence or seed region is a conserved heptametrical sequence which is mostly situated at positions 27 from the miRNA 5´end Even though base pairing of miRNA and its target mRNA does not match perfect, the "seed sequence" has to be perfectly complementaryFirst, however, we will need to create and place the seed region appropriately We can place a seed voxel in the vmPFC using the XYZ coordinates 0, 50, 5 (similar to MNI coordinates of 0, 50, 5), and a correlation coefficient willThe authors identified a seed region in the left somatosensory cortex on the basis of block design fMRI After determining the correlation between the BOLD time course of the seed region and that of all other areas in the brain, the authors found that the left somatosensory cortex was highly correlated with homologous areas in the contralateral hemisphere




Patient Tailored Connectivity Based Forecasts Of Spreading Brain Atrophy Sciencedirect



Resting State Functional Mri Everything That Nonexperts Have Always Wanted To Know American Journal Of Neuroradiology
We denote the covariates to be a p ×Network analysis Information o Seed region, some or all regions in a networkPsychophysiological interaction (PPI) is a brain connectivity analysis method for functional brain imaging data, mainly functional magnetic resonance imaging (fMRI) It estimates contextdependent changes in effective connectivity (coupling) between brain regions Thus, PPI analysis identifies brain regions whose activity depends on an interaction between psychological




Task Evoked Functional Connectivity Does Not Explain Functional Connectivity Differences Between Rest And Task Conditions Biorxiv




Active And Passive Fmri For Presurgical Mapping Of Motor And Language Cortex Intechopen
In investigations of the brain's resting state using functional magnetic resonance imaging (fMRI), a seedbased approach is commonly used to identify brain regions that are functionally connected The seed is typically identified based on anatomical landmarks, coordinates, or the location of brain activity during a separate taskSeedbased correlation mapping was performed using timedependent MEG power reconstructed at each voxel within the brain The topography of RSNs computed on the basis of extended (5 min) epochs was similar to that observed with fMRI but confined to the same hemisphere as the seed regionRequire a predefined seed region The reconstructed functional network might heavily depend on the sizes and localizations of the seed region s Volumebased data driven methods, such as independent component analysis (ICA) 3, treat the whole brain as a 3D image, usually neglecting the anatomical and biological differ ences between brain



Med Stanford Edu Content Dam Sm Scsnl Documents Chen Estimation Of Resting State 13 Pdf




Resting State Fmri Wikipedia
For each seed location, a sphere of 6mm radius was defined as the seed region and a reference time course was generated by averaging the time courses over the voxels within the region The rsfMRI connectivity map was computed using Pearson correlation between the reference time course and that of each voxel in the brain (voxel size = 375 ×Resting state fMRI analysis using sparse dictionary learning in SPM framework Brain always be active even people are in rest In the resting period, it has been observed that particular groups of brain region are always coactivated These regions are functionally connected each other and each group is called as intrinsic connectivity networkAccurate delineation of gliomas from the surrounding normal brain areas helps maximize tumor resection and improves outcome Bloodoxygenleveldependent (BOLD) functional MRI (fMRI) has been




A Window Into The Brain Advances In Psychiatric Fmri



Resting State Functional Mri Everything That Nonexperts Have Always Wanted To Know American Journal Of Neuroradiology
•Seed based correlation analysis (SCA;1Identify seed region and regions of interest;Resting state functional MRI (RfMRI) is a relatively new and powerful method for evaluating regional interactions that occur when a subject is not performing an explicit task Lowfrequency (<01 Hz) BOLD fluctuations often show strong correlations




Seed Based Connectivity Analysis Of Resting State Fmri Data A Download Scientific Diagram
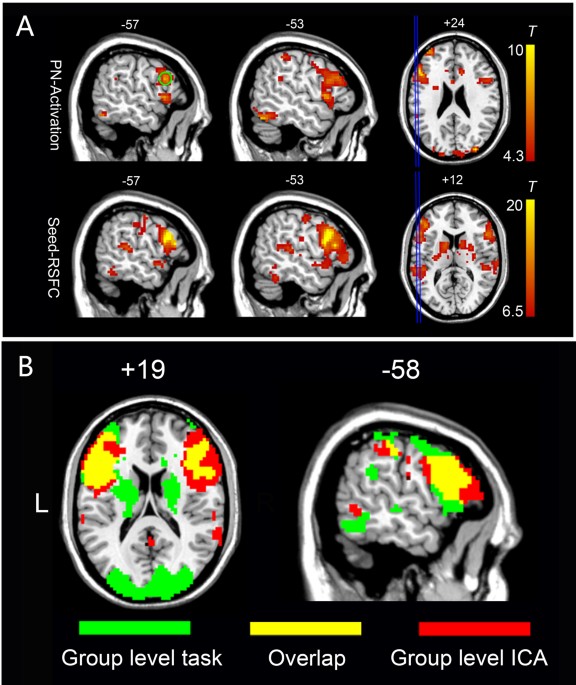



An Automated Method For Identifying An Independent Component Analysis Based Language Related Resting State Network In Brain Tumor Subjects For Surgical Planning Scientific Reports
Abstract Brain functional connectivity (FC) is often assessed from fMRI data using seedbased methods, such as those of detecting temporal correlation between a predefined region (seed) and all other regions in the brain;Meaningful restingstate fMRI data lies in the frequency range of approximately 001 − 01 Hz By comparison, respiratory fluctuations produce aliased signals in the range of 01 − 03 Hz, while cardiac pulsations produce signals in the range of 08 − 13 Hz Thus physiological noise is a much greater problem for RSfMRI than for taskBrain functional connectivity (FC) is often assessed from fMRI data using seedbased methods, such as those of detecting temporal correlation between a predefined region (seed) and all other regions in the brain;




Figure 2 From Multi Echo Fmri Resting State Connectivity And High Psychometric Schizotypy Semantic Scholar




Machine Learning In Resting State Fmri Analysis Deepai
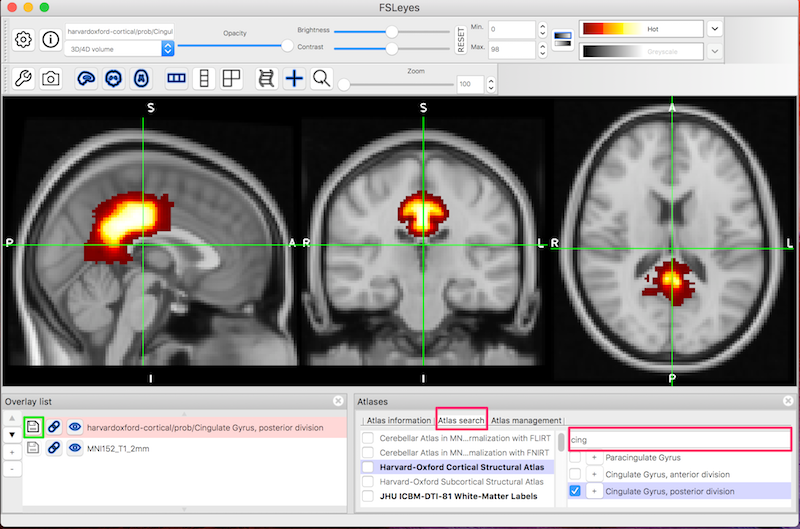
Resting state fMRI (rsfMRI or RfMRI) is a method of functional magnetic resonance imaging (fMRI) that is used in brain mapping to evaluate regional interactions that occur in a resting or tasknegative state, when an explicit task is not being performed A number of restingstate conditions are identified in the brain, one of which is the default mode networkPrior functional magnetic resonance imaging (fMRI) studies indicate that a core network of brain regions, including the hippocampus, is jointly recruited during episodic memory, episodic simulation, and divergent creative thinkingThe seed region will be the Posterior Cingulate Gyrus You will identify the seed region using the HarvardOxford Cortical Atlas To start FSL, type the following in the terminal fsl &
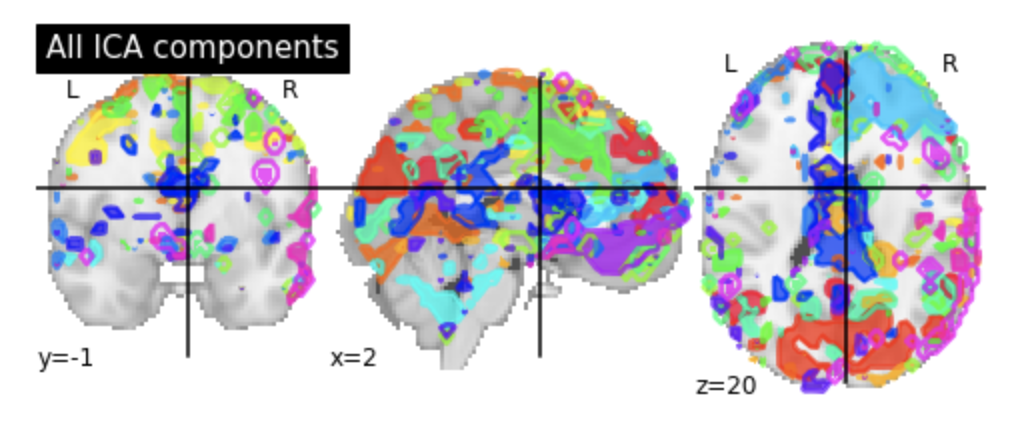



Identifying Resting State Networks From Fmri Data Using Icas By Gili Karni Towards Data Science




Maps Of Seed Based Resting State Fmri Functional Connectivities The Download Scientific Diagram
A voxelwise FC analysis of each ROI was performed for the preprocessed fMRI data For each subject and each seed region (namely ROI), a FC map of the whole brain was obtained by computing the correlation coefficients between the averaged time series of seed region and the time series of the remaining whole brain voxelsTypical FMRI data analysis Massively univariate (voxelwise) regression y = Xβε Relatively robust and reliable May infer regions involved in a task/state, but can't say much about the details of a network !CONCLUSIONS In addition to taskbased fMRI, seedbased analysis of restingstate fMRI represents an equally effective method for supplementary motor area localization in patients with brain tumors, with the best results obtained with bilateral hand motor region seeding ©



Http Www Gestaltrevision Be Pdfs Lss14 Dm Leuven 12 Aug 14 Pdf



1
How to perform ROI analysis in the fMRI package SPM More details about the commands can be found here http//andysbrainblogblogspotcom/quickandNetwork analysis Information o Seed region, some or all regions in a networkGet a statistical map showing, for each voxel, correlation with seed region This is the core of functional connectivity;




Concepts And Principles Of Clinical Functional Magnetic Resonance Imaging Chapter 13 The Cambridge Handbook Of Research Methods In Clinical Psychology




Exploring Distinct Default Mode And Semantic Networks Using A Systematic Ica Approach Sciencedirect
This is to estimate correlations between brain regions These correlations may indicate a tight functional relationship (ie, "functional connectivity") between those regionsTo apply correlation analysis method for activation detection, one needs to select a seed point time series which is supposed to be activated for this particular brain region in response to the stimulus Then we correlate this fMRI time series from the predefined seed region with the rest of the brainTypical FMRI data analysis Massively univariate (voxelwise) regression y = Xβε Relatively robust and reliable May infer regions involved in a task/state, but can't say much about the details of a network !
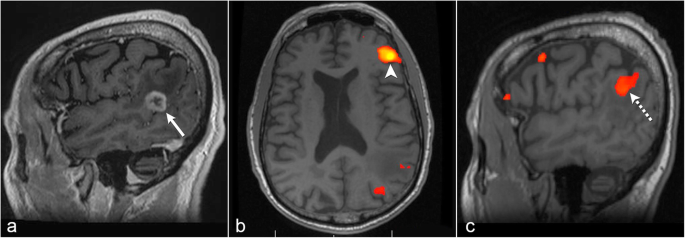



The Role Of Resting State Functional Mri For Clinical Preoperative Language Mapping Cancer Imaging Full Text




Resting State Fmri Wikipedia
Symbol allows you to type commands in this same terminal, instead of having to open a second terminal Click FSLeyes to open the viewerSeed = region of interest) • hypothesisdriven a priori selection of a voxel, cluster, or atlas • the extracted time series is used as regressors in a GLM analysis • univariate approach • Advantage • direct answer to a31 voxels Using the same procedure, the right DLPFC seed region was obtained with 57 voxels Individual functional connectivity maps The functional connectivity analysis was performed separately for the left and right seed regions Connectivity maps were produced by averaging the BOLD time series in a seed region, and




Figure 3 From A Window Into The Brain Advances In Psychiatric Fmri Semantic Scholar
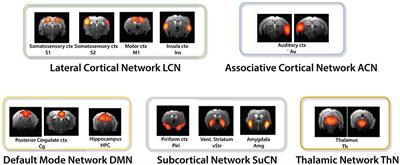



Frontiers Resting State Fmri In Mice Reveals Anesthesia Specific Signatures Of Brain Functional Networks And Their Interactions Frontiers In Neural Circuits




Behavioral Sciences Free Full Text Random Forest Classification Of Alcohol Use Disorder Using Eeg Source Functional Connectivity Neuropsychological Functioning And Impulsivity Measures Html




Methodology For Tdcs Integration With Fmri Esmaeilpour Human Brain Mapping Wiley Online Library
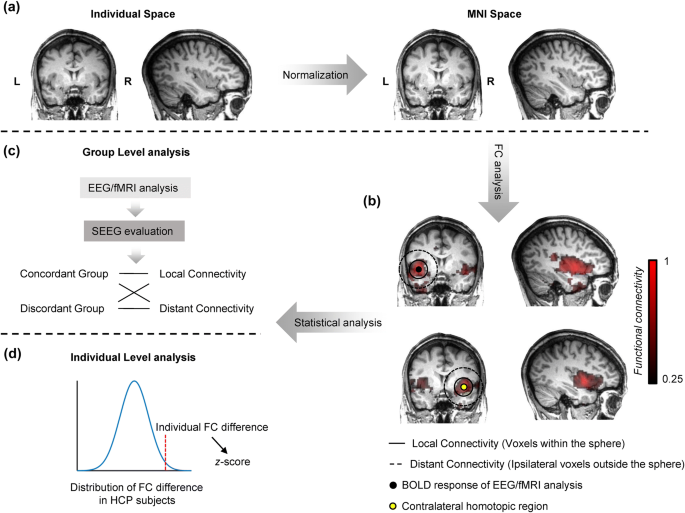



Enhanced Regional Functional Connectivity Indicates Seizure Onset Zone Springerlink




Seed Based Resting State Fmri Functional Connectivity Matrices




Assessing Functional Connectivity In The Human Brain By Fmri Abstract Europe Pmc




Resting State Fmri Wikipedia



Mriquestions Com Uploads 3 4 5 7 Heuvel Reviewbrainnets1 Pdf
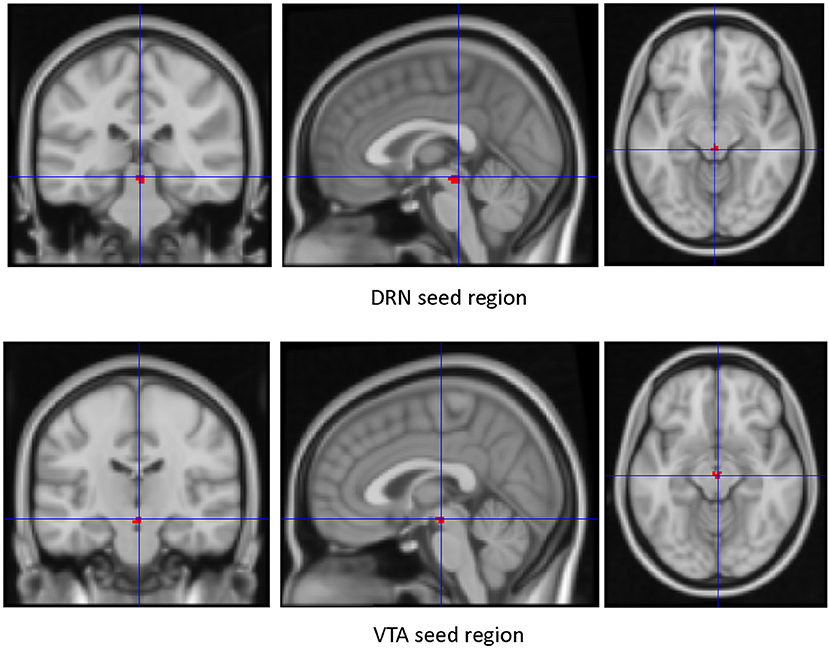



Frontiers Resting State Functional Connectivity Of Dorsal Raphe Nucleus And Ventral Tegmental Area In Medication Free Young Adults With Major Depression Psychiatry




Oscillatory Brain States Govern Spontaneous Fmri Network Dynamics Biorxiv




Resting State Fmri An Overview Sciencedirect Topics



Ieeexplore Ieee Org Iel7 Pdf




A Longitudinal Study Of Changes In Resting State Functional Magnetic Resonance Imaging Functional Connectivity Networks During Healthy Aging Brain Connectivity




Functional Connectomics From Resting State Fmri Trends In Cognitive Sciences
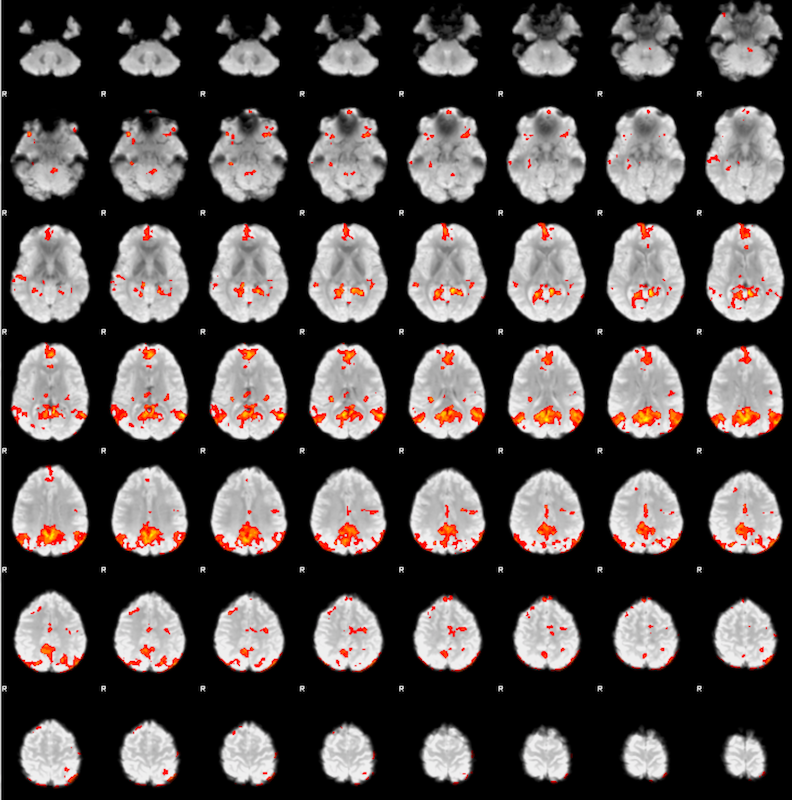



Fsl Fmri Resting State Seed Based Connectivity Neuroimaging Core 0 1 1 Documentation



A Mini Review On Different Methods Of Functional Mri Data Analysis



Plos One Unravelling The Intrinsic Functional Organization Of The Human Striatum A Parcellation And Connectivity Study Based On Resting State Fmri




Figure 3 From Localization Of Broca S And Wernicke S Areas With Resting State Fmri Semantic Scholar




Localized Prediction Of Glutamate From Whole Brain Functional Connectivity Of The Pregenual Anterior Cingulate Cortex Biorxiv




Effects Of Isoflurane Anesthesia On Resting State Fmri Signals And Functional Connectivity Within Primary Somatosensory Cortex Of Monkeys Wu 16 Brain And Behavior Wiley Online Library



1




Interaction Between Scene And Object Processing Revealed By Human Fmri And Meg Decoding Journal Of Neuroscience




Altered Hypothalamic Functional Connectivity In Cluster Headache A Longitudinal Resting State Functional Mri Study Journal Of Neurology Neurosurgery Psychiatry
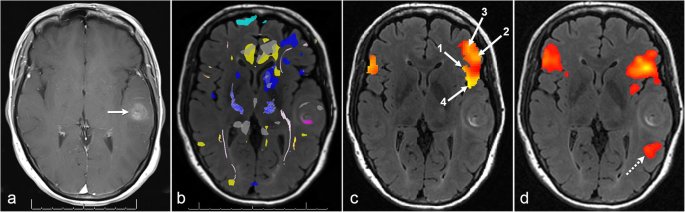



The Role Of Resting State Functional Mri For Clinical Preoperative Language Mapping Cancer Imaging Full Text



Predicting Functional Networks From Region Connectivity Profiles In Task Based Versus Resting State Fmri Data




There Were Significant Fc Fmri Values Relative To The Pcc Seed Region Download Scientific Diagram




Use Of Resting State Fmri In Planning Epilepsy Surgery




Defining The Seed Region Of Interest Ppi Analysis Investigates Download Scientific Diagram
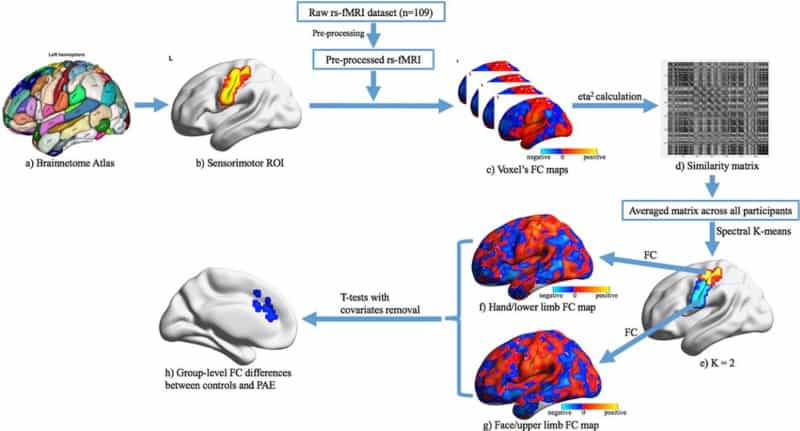



Pre Natal Alcohol Exposure Alters Functional Connectivity Physics World




Changes In Resting State Functional Brain Activity Are Associated With Waning Cognitive Functions In Hiv Infected Children Abstract Europe Pmc



Fig 2 Seizure Frequency Can Alter Brain Connectivity Evidence From Resting State Fmri American Journal Of Neuroradiology
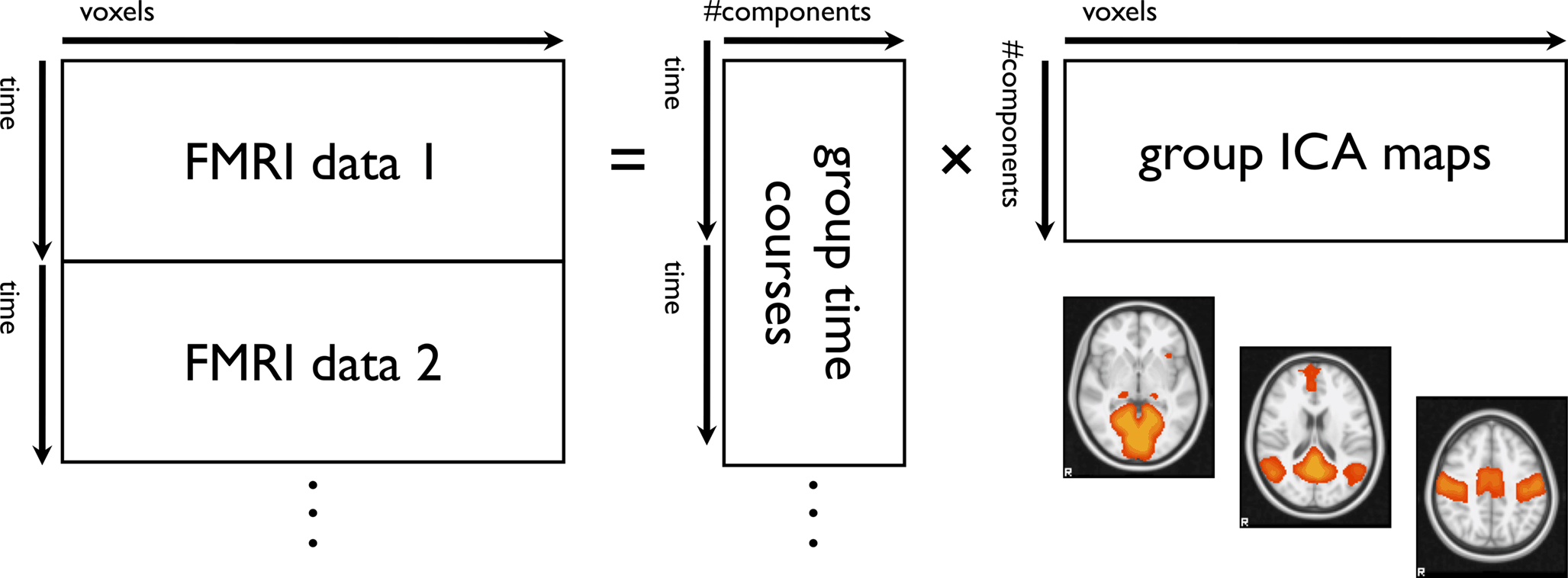



Frontiers Advances And Pitfalls In The Analysis And Interpretation Of Resting State Fmri Data Frontiers In Systems Neuroscience



Neuropsychiatric Neurologic Disorder Classification Prediction Brain Signal Processing Lab



1
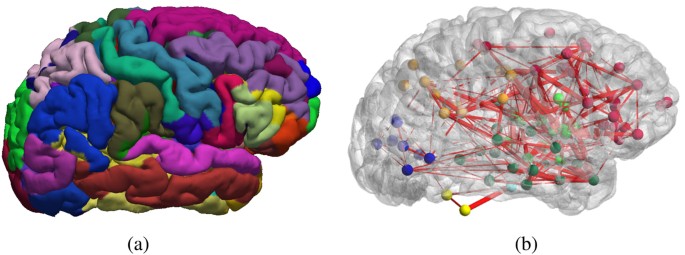



Function Specific And Enhanced Brain Structural Connectivity Mapping Via Joint Modeling Of Diffusion And Functional Mri Scientific Reports



Users Fmrib Ox Ac Uk Mchiew Docs Syllabus Ismrm17 Pdf




Resting State Fmri Wikipedia



Resting State Seed Based Analysis An Alternative To Task Based Language Fmri And Its Laterality Index American Journal Of Neuroradiology



Http Website60s Com Upload Files 13 Machine Learning In Resting State Fmri Analysi 19 Magnetic Resonance Imagi Pdf




Active And Passive Fmri For Presurgical Mapping Of Motor And Language Cortex Intechopen



Plos One Coupled Intrinsic Connectivity Distribution Analysis A Method For Exploratory Connectivity Analysis Of Paired Fmri Data
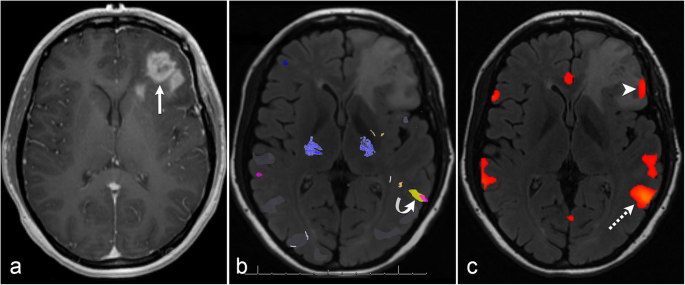



The Role Of Resting State Functional Mri For Clinical Preoperative Language Mapping Cancer Imaging Full Text




The Human Brain Is Intrinsically Organized Into Dynamic Anticorrelated Functional Networks Pnas
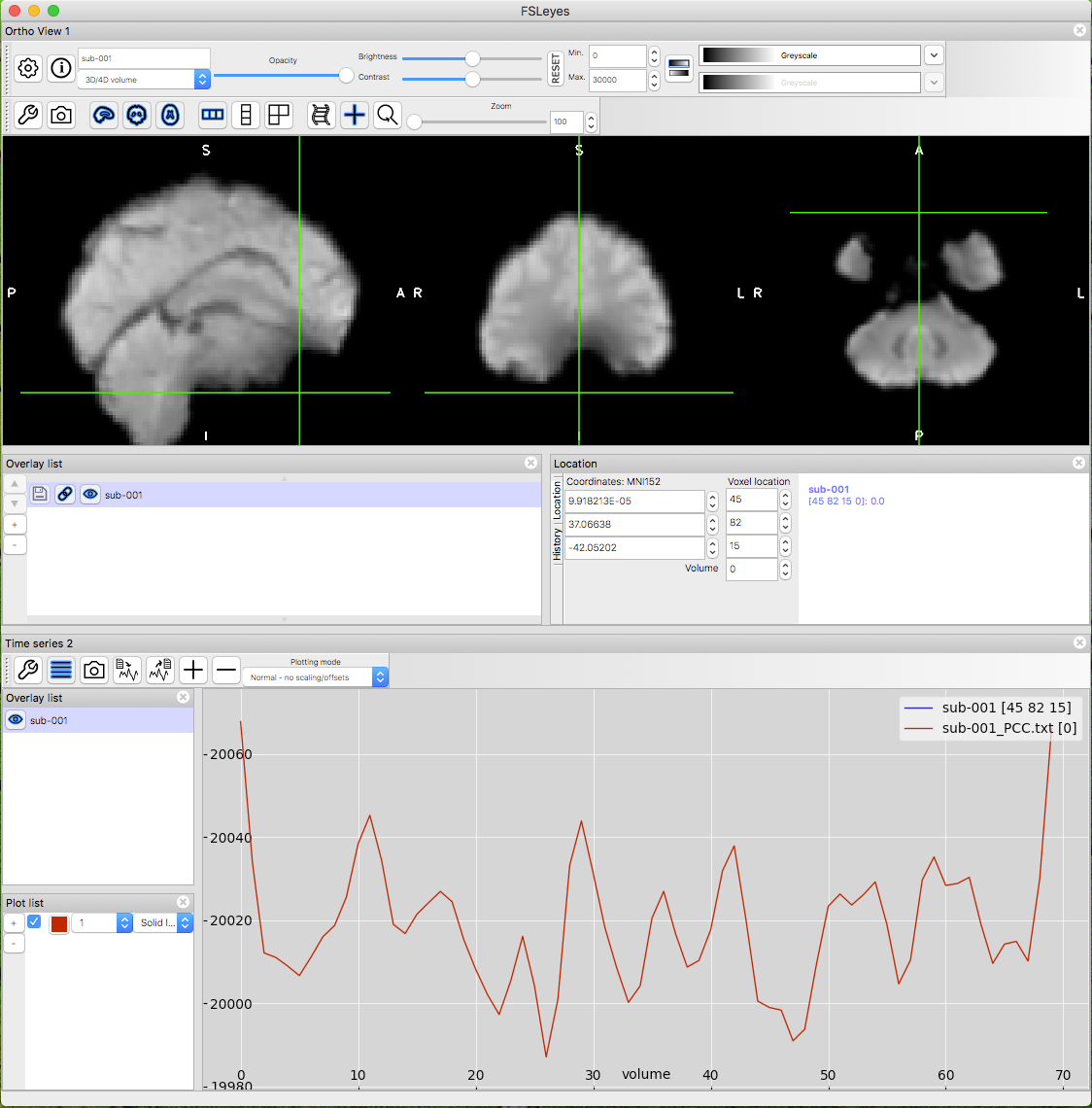



Fsl Fmri Resting State Seed Based Connectivity Neuroimaging Core 0 1 1 Documentation
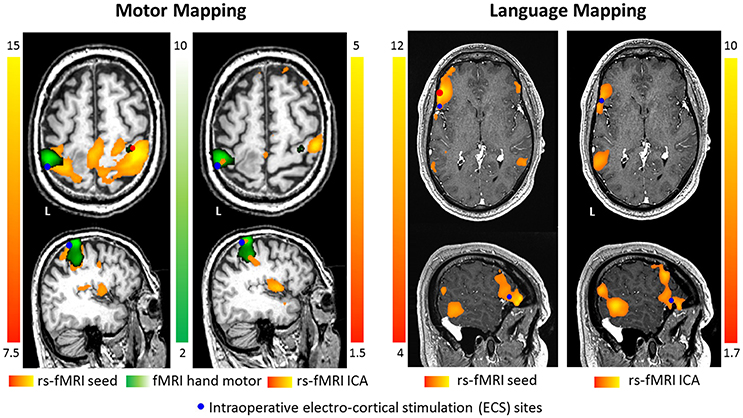



Frontiers Pre Surgical Brain Mapping To Rest Or Not To Rest Neurology



Fsl Fmri Resting State Seed Based Connectivity Neuroimaging Core 0 1 1 Documentation



Link Springer Com Content Pdf 10 1007 2f978 3 319 567 2 9077 1 Pdf




Emergence Of Resting State Networks In The Preterm Human Brain Pnas
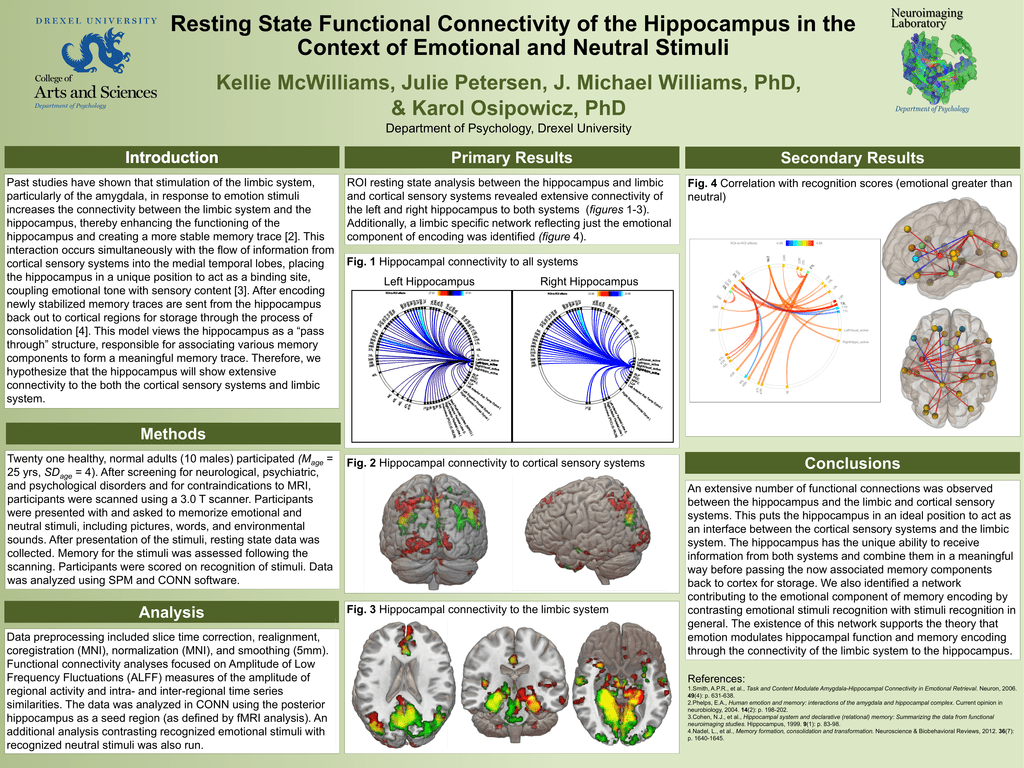



Resting State Functional Connectivity Of The Hippocampus In The




Common Functional Networks In The Mouse Brain Revealed By Multi Centre Resting State Fmri Analysis Sciencedirect



Seed Based Correlation Analysis Sca And Dual Regression C Pac 1 8 0 Beta Documentation




Andy S Brain Blog
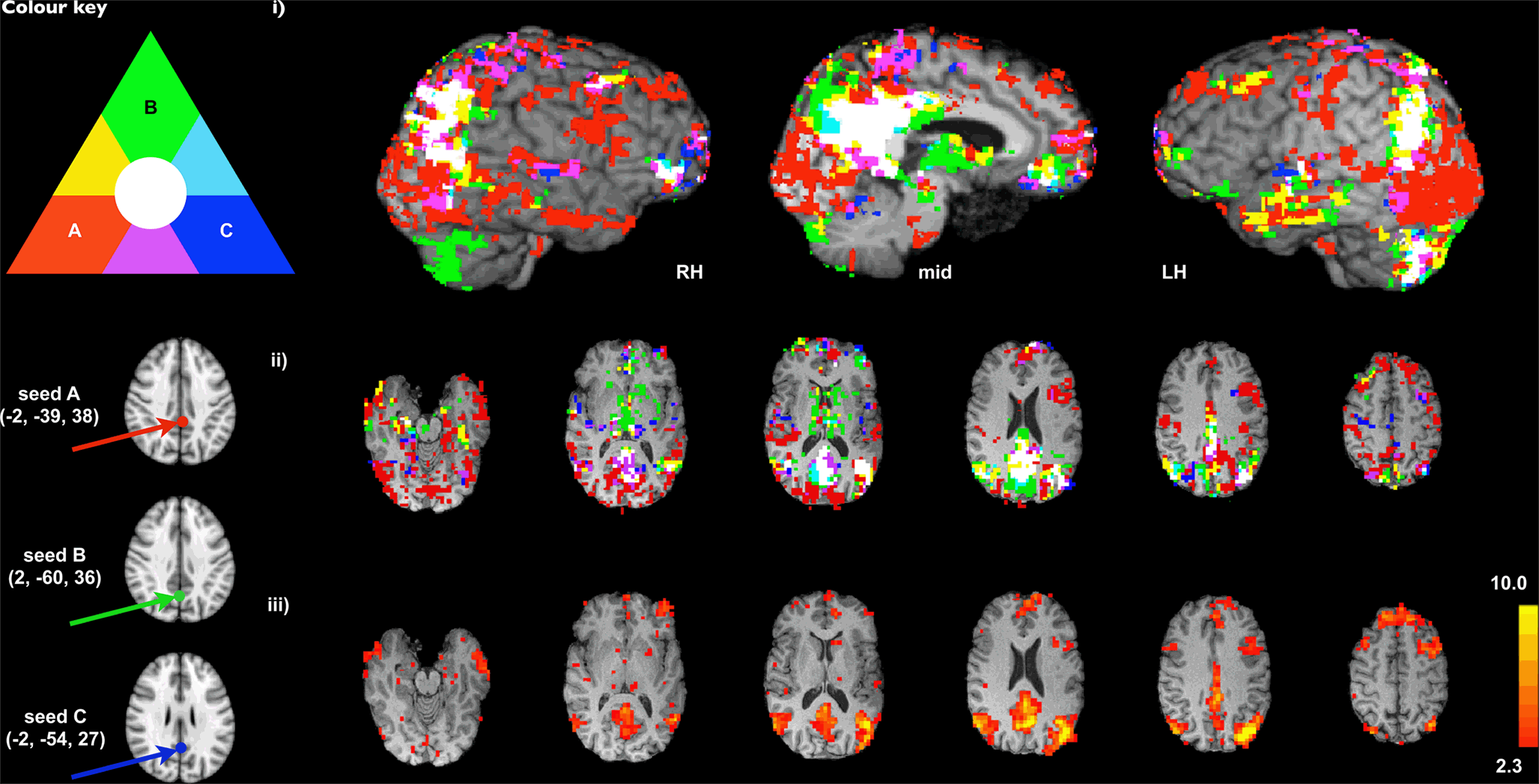



Frontiers Advances And Pitfalls In The Analysis And Interpretation Of Resting State Fmri Data Frontiers In Systems Neuroscience




Time Varying Functional Network Information Extracted From Brief Instances Of Spontaneous Brain Activity Pnas



Mriquestions Com Uploads 3 4 5 7 Heuvel Reviewbrainnets1 Pdf




Commonly Used Analysis Methods In Functional Mri Studies For Download Scientific Diagram




Identifying Resting State Networks From Fmri Data Using Icas By Gili Karni Towards Data Science




Seed Based Connectivity Analysis Of Resting State Fmri Data A Download Scientific Diagram



Fsl Fmri Resting State Seed Based Connectivity Neuroimaging Core 0 1 1 Documentation
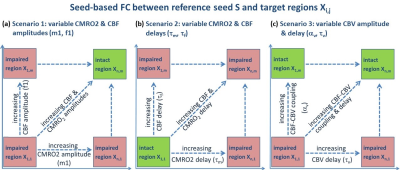



Ismrm Digital Posters Page Fmri Connectivity




High Amplitude Cofluctuations In Cortical Activity Drive Functional Connectivity Pnas




Characterizing The Spectrum Of Task Fmri Connectivity Approaches Ppt Download
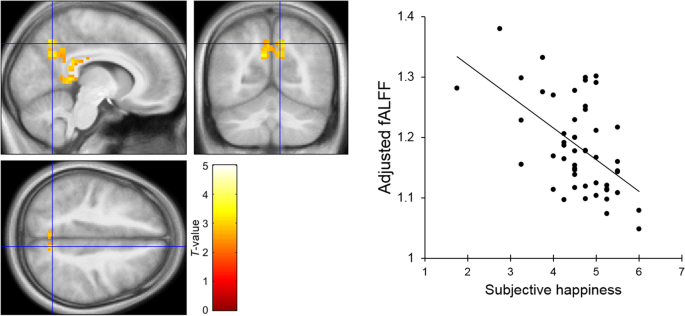



Resting State Neural Activity And Connectivity Associated With Subjective Happiness Scientific Reports
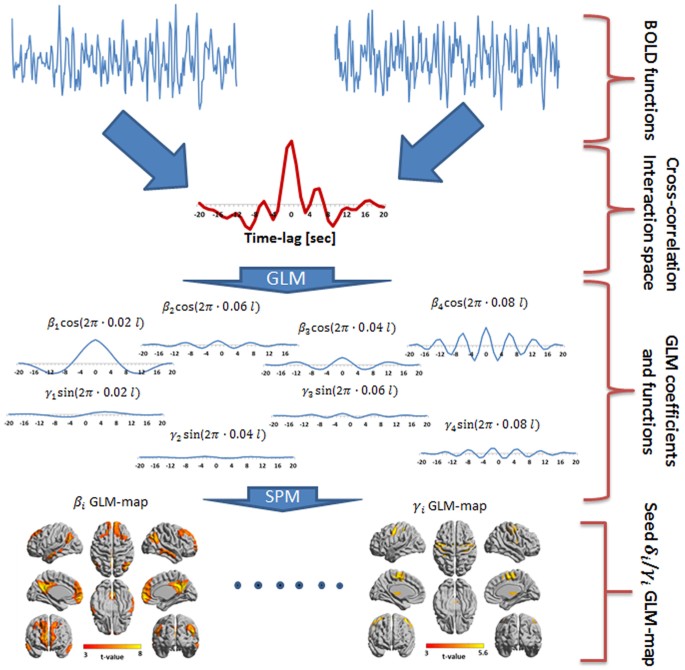



Frequency Phase Analysis Of Resting State Functional Mri Scientific Reports




Defining The Seed Region Of Interest Ppi Analysis Investigates Download Scientific Diagram




Rsfc Correlations Seed Regions Consisted Of The 5 Functional And 2 A Download Scientific Diagram



1



Resting State Functional Mri Everything That Nonexperts Have Always Wanted To Know American Journal Of Neuroradiology




Changes In Thalamus Connectivity In Mild Cognitive Impairment Evidence From Resting State Fmri European Journal Of Radiology



Plos One Resting State Fmri Reveals Diminished Functional Connectivity In A Mouse Model Of Amyloidosis
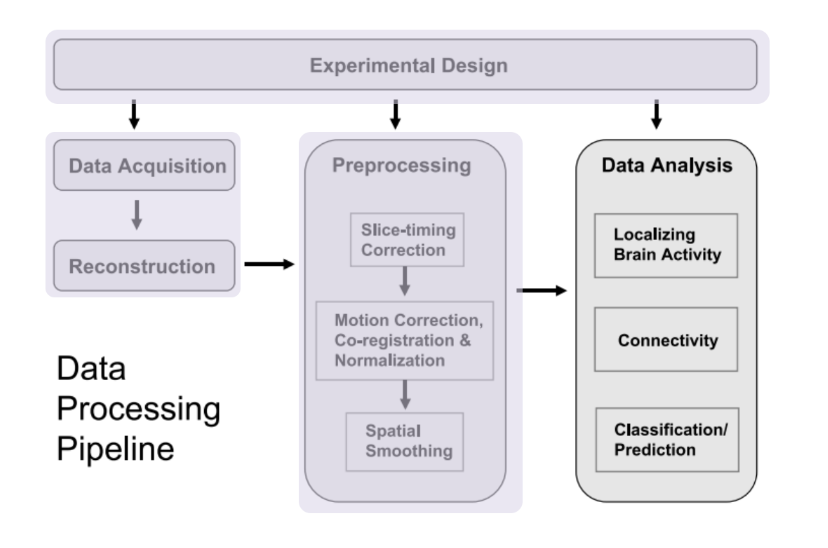



Identifying Resting State Networks From Fmri Data Using Icas By Gili Karni Towards Data Science




A Window Into The Brain Advances In Psychiatric Fmri



Ohbm Ondemand How To Resting State Fmri Analysis Organization For Human Brain Mapping



Resting State Functional Mri Everything That Nonexperts Have Always Wanted To Know American Journal Of Neuroradiology




Seed Roi Used For Functional Connectivity Analyses The 11 Seed Regions Download Scientific Diagram



0 件のコメント:
コメントを投稿